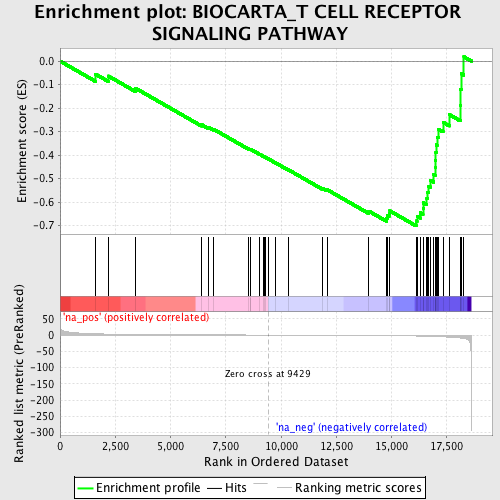
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
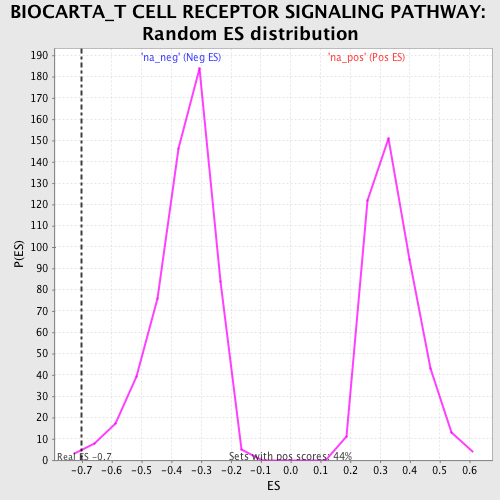
| Enrichment Score (ES) | -0.70136464 |
| Normalized Enrichment Score (NES) | -1.9395174 |
| Nominal p-value | 0.0053380784 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.865 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTPN7 | 1600 | 4.062 | -0.0552 | No | ||
| 2 | MAPK8 | 2179 | 3.027 | -0.0632 | No | ||
| 3 | MAP2K1 | 3408 | 1.872 | -0.1150 | No | ||
| 4 | CD4 | 6400 | 0.764 | -0.2703 | No | ||
| 5 | ARHGAP5 | 6737 | 0.677 | -0.2832 | No | ||
| 6 | CD247 | 6934 | 0.624 | -0.2890 | No | ||
| 7 | CD3E | 8523 | 0.227 | -0.3728 | No | ||
| 8 | RALBP1 | 8611 | 0.207 | -0.3759 | No | ||
| 9 | CD3G | 8635 | 0.200 | -0.3756 | No | ||
| 10 | FOS | 9018 | 0.107 | -0.3953 | No | ||
| 11 | HRAS | 9216 | 0.053 | -0.4055 | No | ||
| 12 | CAMK2B | 9238 | 0.048 | -0.4063 | No | ||
| 13 | CD3D | 9273 | 0.038 | -0.4078 | No | ||
| 14 | LAT | 9409 | 0.005 | -0.4151 | No | ||
| 15 | PLCG1 | 9738 | -0.081 | -0.4321 | No | ||
| 16 | SOS1 | 10324 | -0.226 | -0.4619 | No | ||
| 17 | PIK3R1 | 11865 | -0.608 | -0.5402 | No | ||
| 18 | TRB@ | 12085 | -0.673 | -0.5468 | No | ||
| 19 | PPP3CA | 13975 | -1.312 | -0.6385 | No | ||
| 20 | ARHGAP6 | 14779 | -1.669 | -0.6690 | No | ||
| 21 | RAF1 | 14806 | -1.685 | -0.6576 | No | ||
| 22 | SHC1 | 14889 | -1.736 | -0.6487 | No | ||
| 23 | RAC1 | 14926 | -1.756 | -0.6373 | No | ||
| 24 | ZAP70 | 16117 | -2.799 | -0.6800 | Yes | ||
| 25 | ARHGAP1 | 16157 | -2.853 | -0.6603 | Yes | ||
| 26 | FYN | 16296 | -3.029 | -0.6447 | Yes | ||
| 27 | PIK3CA | 16426 | -3.246 | -0.6269 | Yes | ||
| 28 | PTPRC | 16439 | -3.278 | -0.6025 | Yes | ||
| 29 | ARFGAP1 | 16598 | -3.555 | -0.5839 | Yes | ||
| 30 | GRB2 | 16640 | -3.636 | -0.5584 | Yes | ||
| 31 | ARFGAP3 | 16687 | -3.718 | -0.5325 | Yes | ||
| 32 | PPP3CB | 16752 | -3.824 | -0.5067 | Yes | ||
| 33 | ARHGAP4 | 16904 | -4.143 | -0.4833 | Yes | ||
| 34 | PPP3CC | 16975 | -4.313 | -0.4541 | Yes | ||
| 35 | LCK | 16984 | -4.337 | -0.4215 | Yes | ||
| 36 | MAP2K4 | 17006 | -4.402 | -0.3890 | Yes | ||
| 37 | MAP3K1 | 17023 | -4.458 | -0.3559 | Yes | ||
| 38 | RELA | 17065 | -4.586 | -0.3231 | Yes | ||
| 39 | JUN | 17135 | -4.762 | -0.2905 | Yes | ||
| 40 | MAPK3 | 17335 | -5.344 | -0.2605 | Yes | ||
| 41 | NFKBIA | 17625 | -6.306 | -0.2279 | Yes | ||
| 42 | NCF2 | 18115 | -8.749 | -0.1875 | Yes | ||
| 43 | VAV1 | 18125 | -8.843 | -0.1205 | Yes | ||
| 44 | PRKCA | 18153 | -9.054 | -0.0529 | Yes | ||
| 45 | NFATC1 | 18277 | -10.197 | 0.0183 | Yes |